MiAIRR-to-NCBI Submission Manual#
Scope of this document#
Provide a user manual describing the submission of AIRR data using the NCBI reference implementation described in [Rubelt_2017]. This implementation uses NCBI’s BioProject, BioSample, Sequence Read Archive (SRA) and GenBank repositories and metadata standards to report AIRR data.
Step 1. MiAIRR data submission to BioProject, BioSample and SRA#
Since we propose to include a combination of raw and processed sequence data, the AIRR standard will sometimes need to be distributed and linked across multiple repositories (e.g., data in SRA linked to related data in GenBank). Besides, the data elements that comprise the standard will be mapped to ontologies in BioPortal through NIH CDE (Common Data Element) terms. These linkages will support more sophisticated validation and logical inference.
There are three main alternatives to submit raw AIRR data/metadata to NCBI repositories: (1) CEDAR’s CAIRR pipeline; (2) NCBI’s Web interface; and (3) NCBI’s FTP server. These alternatives are described below:
Option 1. Submission via the CEDAR system (CAIRR submission pipeline)#
CEDAR’s CAIRR submission pipeline helps investigators and curators to edit and validate ontology-controlled metadata. This pipeline provides a seamless interface to transmit SRA datasets to the NCBI SRA and BioSample repositories from the CEDAR Workbench. The pipeline can be directly be accessed at http://cairr.airr-community.org. Note that the CEDAR template and template elements used by this pipeline are publicly available in the following CEDAR folder: All/Shared/Shared by CEDAR/MiAIRR.
Submission steps:
Open CEDAR’s MiAIRR template by clicking on http://cairr.airr-community.org. If you are not already logged in, this will take you to the CEDAR login panel. If you are a new user, you will have to create an account on the CEDAR Workbench by clicking here.
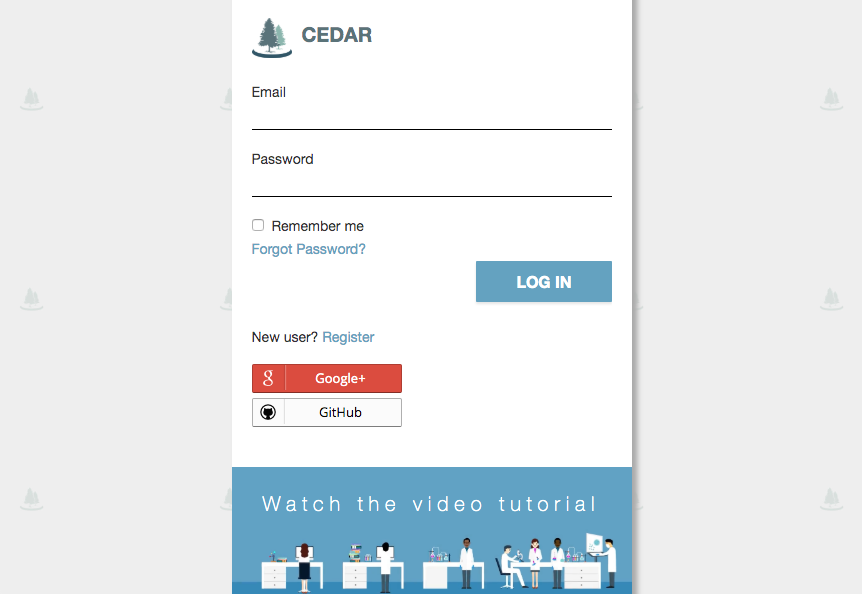
After logging in into the system, you will see the ‘MiAIRR’ template. Fill out the template fields with your metadata. Fields with an asterisk (*) are mandatory. Your submission will fail if any mandatory fields are not completed. If information is unavailable for any mandatory field, please enter ‘not collected’, ‘not applicable’ or ‘missing’ as appropriate. Note that you will need to enter a BioProject ID into the field ‘Study ID’. If you do not have a BioProject yet, you can create one at https://submit.ncbi.nlm.nih.gov/subs/bioproject/
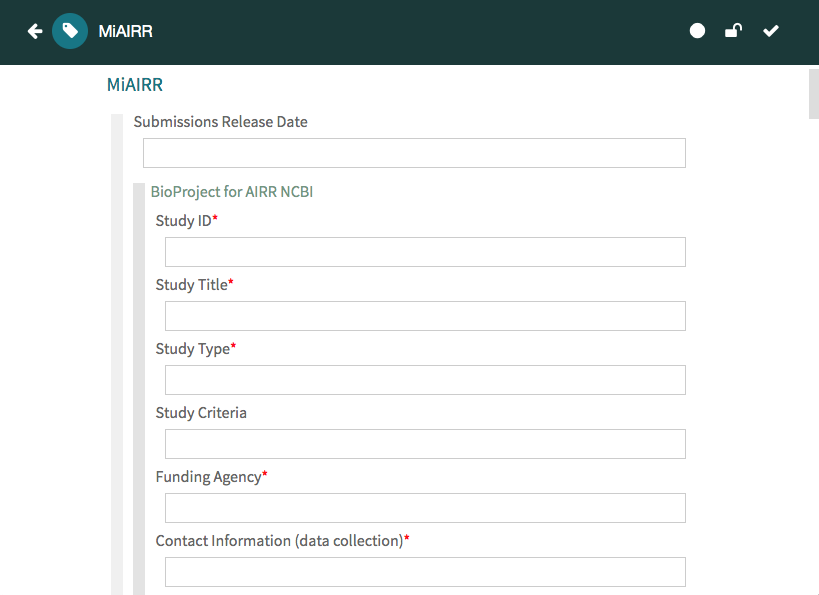
Once your metadata is complete, click on the ‘Save’ button to save your metadata into your workspace. You will see a message in a green box confirming that your metadata have been successfully saved, as well as a message in a yellow box letting you know that your metadata have been saved to your personal workspace.
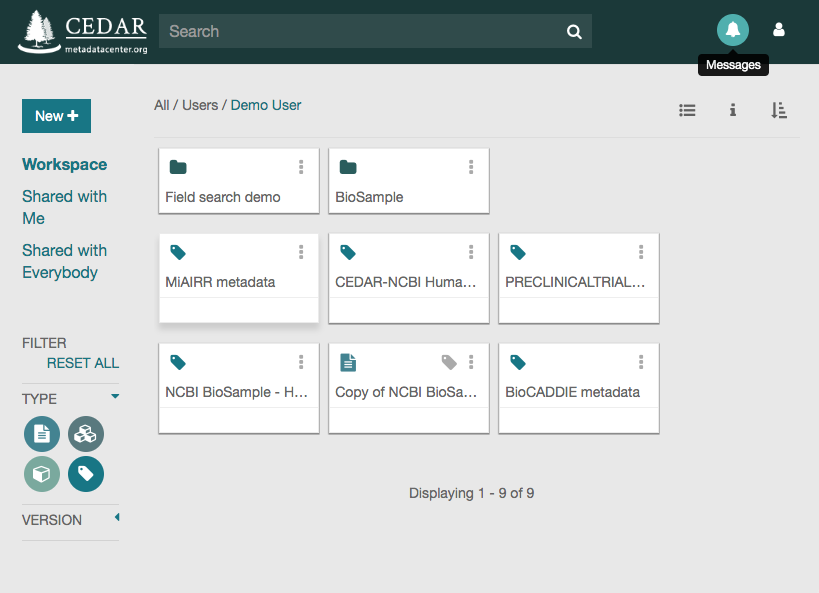
Go to your personal workspace by clicking on the left arrow (top left corner) and then on the ‘Workspace’ link, or by just clicking on: https://cedar.metadatacenter.org
Once in your workspace, you will see a metadata file called ‘MiAIRR metadata’. That file contains the metadata that you have just created and that you want to submit to the NCBI. Click on the three vertical dots on the top-right corner of the file icon to see the available file options.
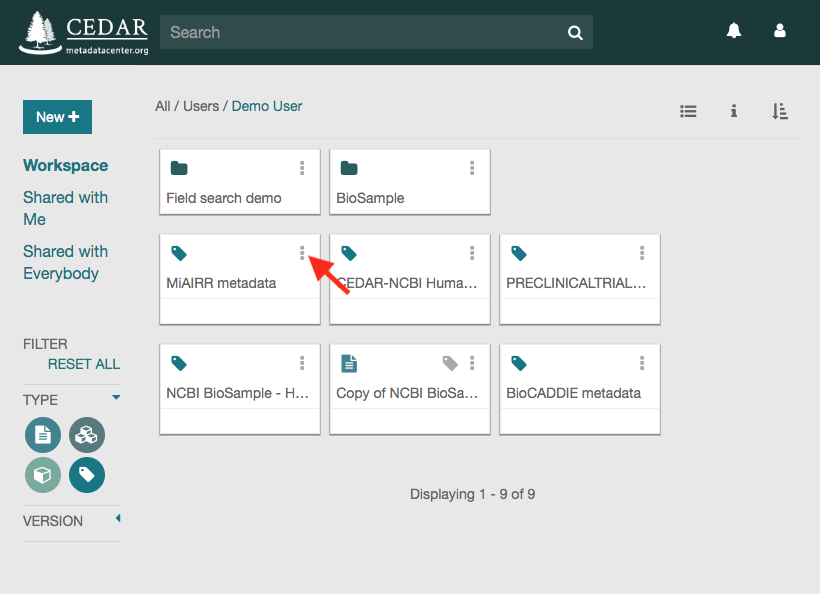
Click on the ‘Submit’ option to open the submission dialog.
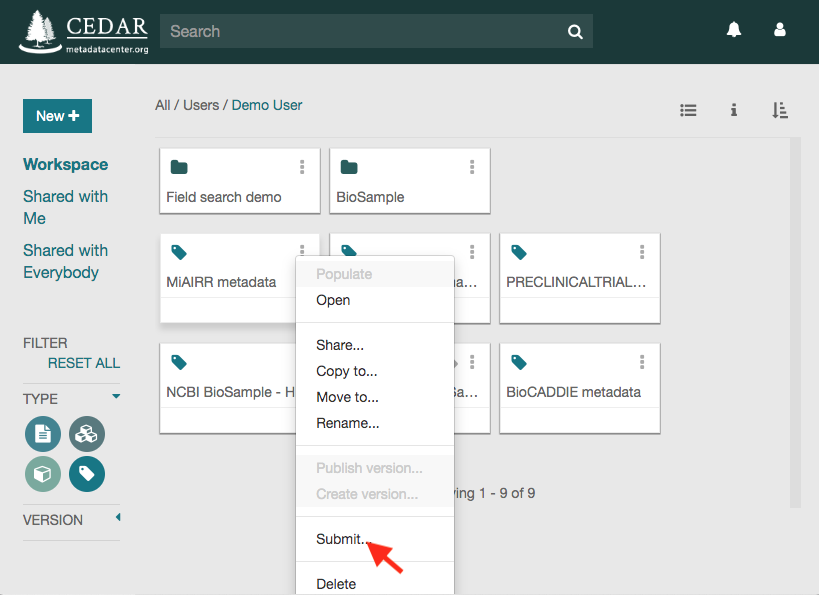
The ‘NCBI MiAIRR’ option will be automatically selected. Click on ‘Next’ to go to the next step.
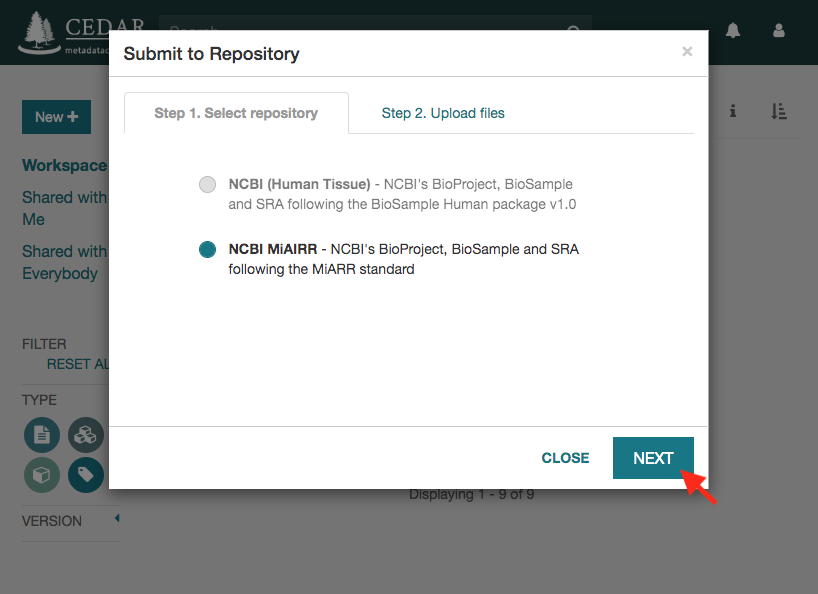
Click on the ‘Select Files’ button to upload the data files. Note that the names of the selected files must match the names in the metadata file. Otherwise, you will receive an error message when trying to start the submission.
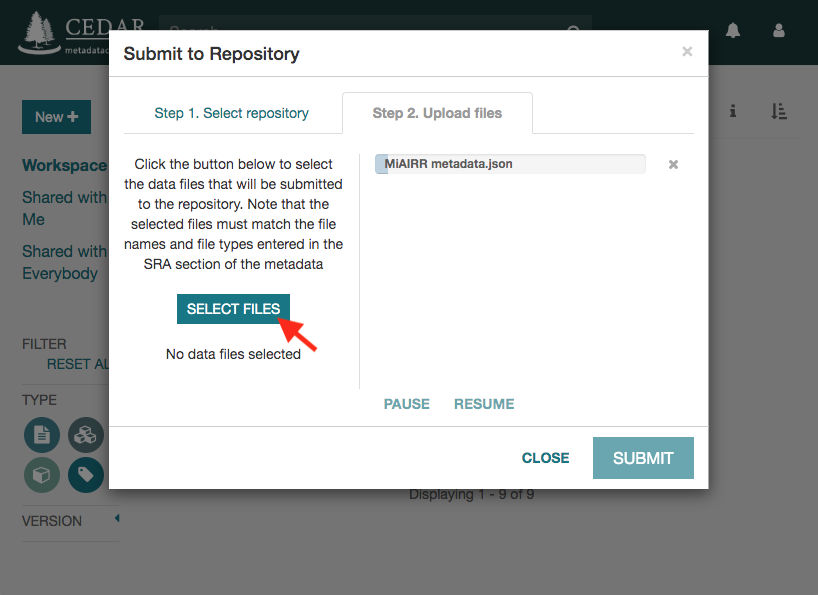
Click on the ‘Submit’ button to start the submission. If there are not validation errors, the selected data files and the corresponding metadata will be uploaded to the NCBI servers.
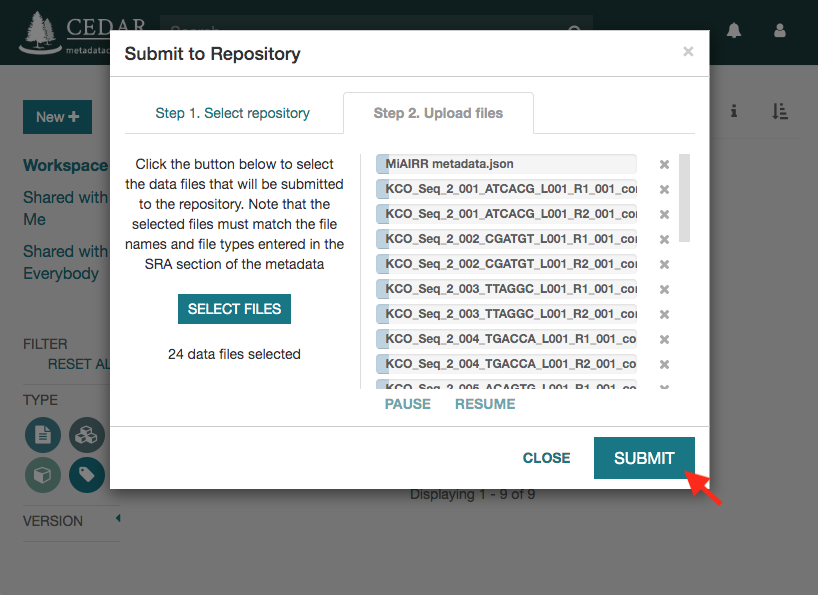
Note that the submission may take several hours or even days to be processed by the NCBI. Meanwhile, you will receive status messages about your submission in your workspace (messages icon).
Proceed with deposit of processed data, below.
Citing the CAIRR pipeline#
Bukhari, Syed Ahmad Chan, Martin J. O’Connor, Marcos Martínez-Romero, Attila L. Egyedi, Debra Debra Willrett, John Graybeal, Mark A. Musen, Florian Rubelt, Kei H. Cheung, and Steven H. Kleinstein. The CAIRR pipeline for submitting standards-compliant B and T cell receptor repertoire sequencing studies to the NCBI. Frontiers in Immunology 9 (2018): 1877. DOI: 10.3389/fimmu.2018.01877
Tell Us About It#
Please let us know how it went! If you are willing, we would love to have your comments in a short survey, it should just take 5 minutes or so. We also welcome entry of issues and requests in our GitHub repository, and emails can be sent to cedar-users@lists.stanford.edu. Both of these resources are publicly visible.
Support or Contact#
Having trouble with NCBI submission process through our pipeline? Please email to Syed Ahmad Chan Bukhari or to Marcos Martínez-Romero and we will help you sort it out.
Option 2. Submission via NCBI’s web interface#
To facilitate AIRR data submissions to NCBI repositories, we have developed the NCBI-compliant metadata submission templates both for single and bulk AIRR data submissions. NCBI provides a web-based interface to create a BioProject and allows to BioSample, Sequence Read Archive (SRA) and GenBank metadata via tab-delimited files for single BioProject related data files submission.
Submitting AIRR data and associated metadata to the Bioproject, BioSample and SRA repositories via NCBI’s web interface follows in general the submission procedure described in [NCBI_NBK47528], but uses AIRR-specific template for metadata submission:
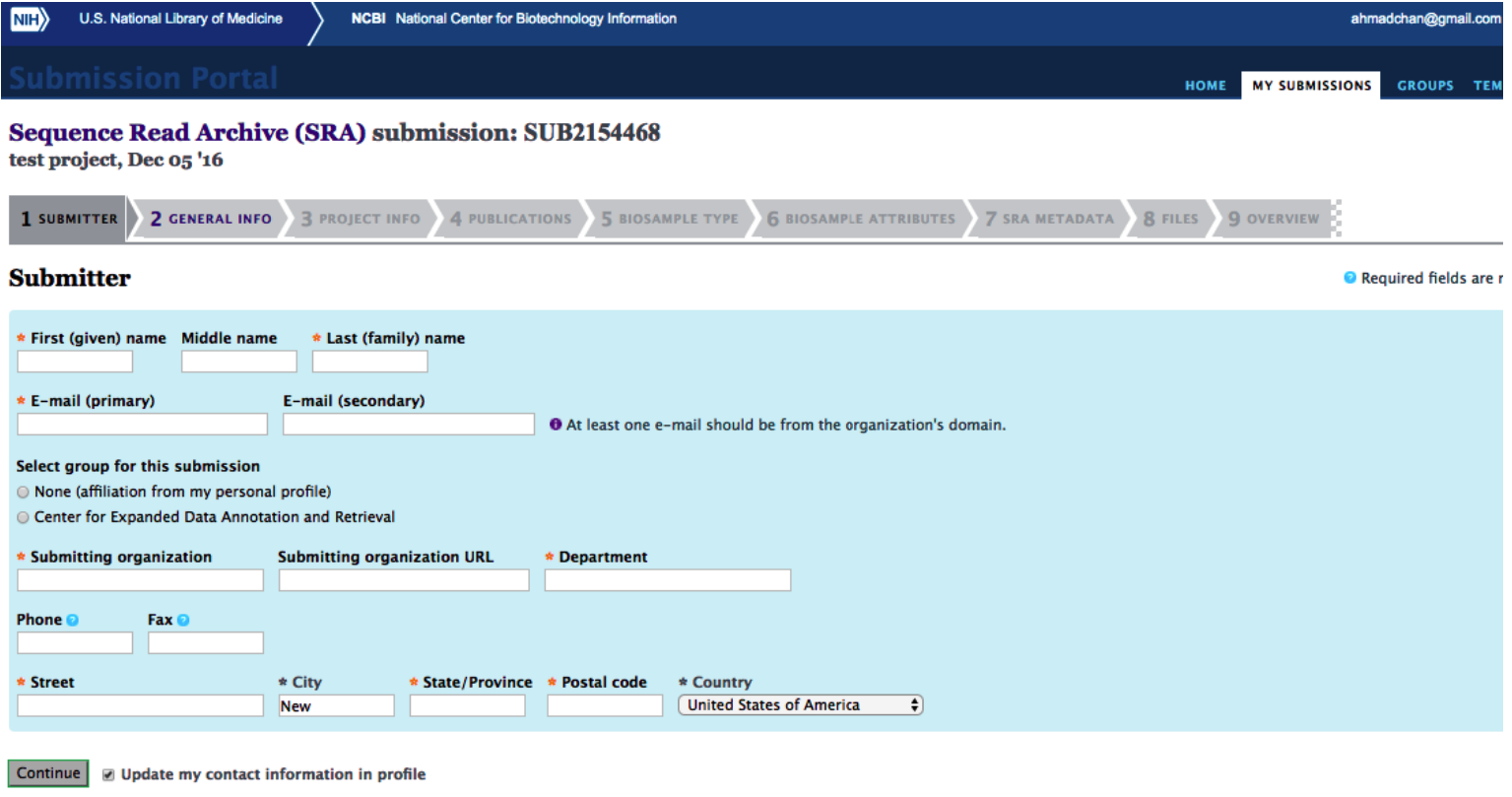
Go to https://submit.ncbi.nlm.nih.gov/subs/sra/ and login with your NCBI account (create an account if necessary).
Click on “create new submission”. You will see a form as below. Fill the form with required information and click on “continue”.
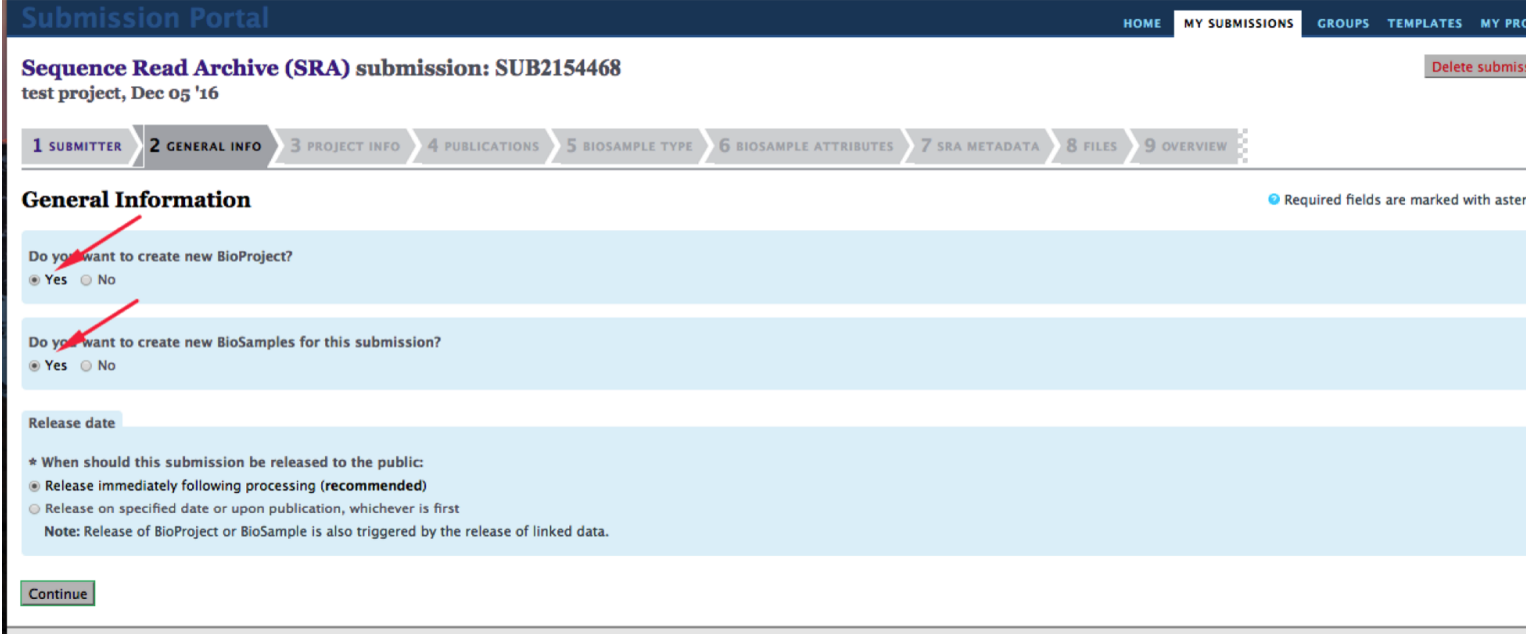
If you are submitting for the first time, check “Yes” on the “new BioProject” or “new BioSample” options to create a new project or sample, respectively.
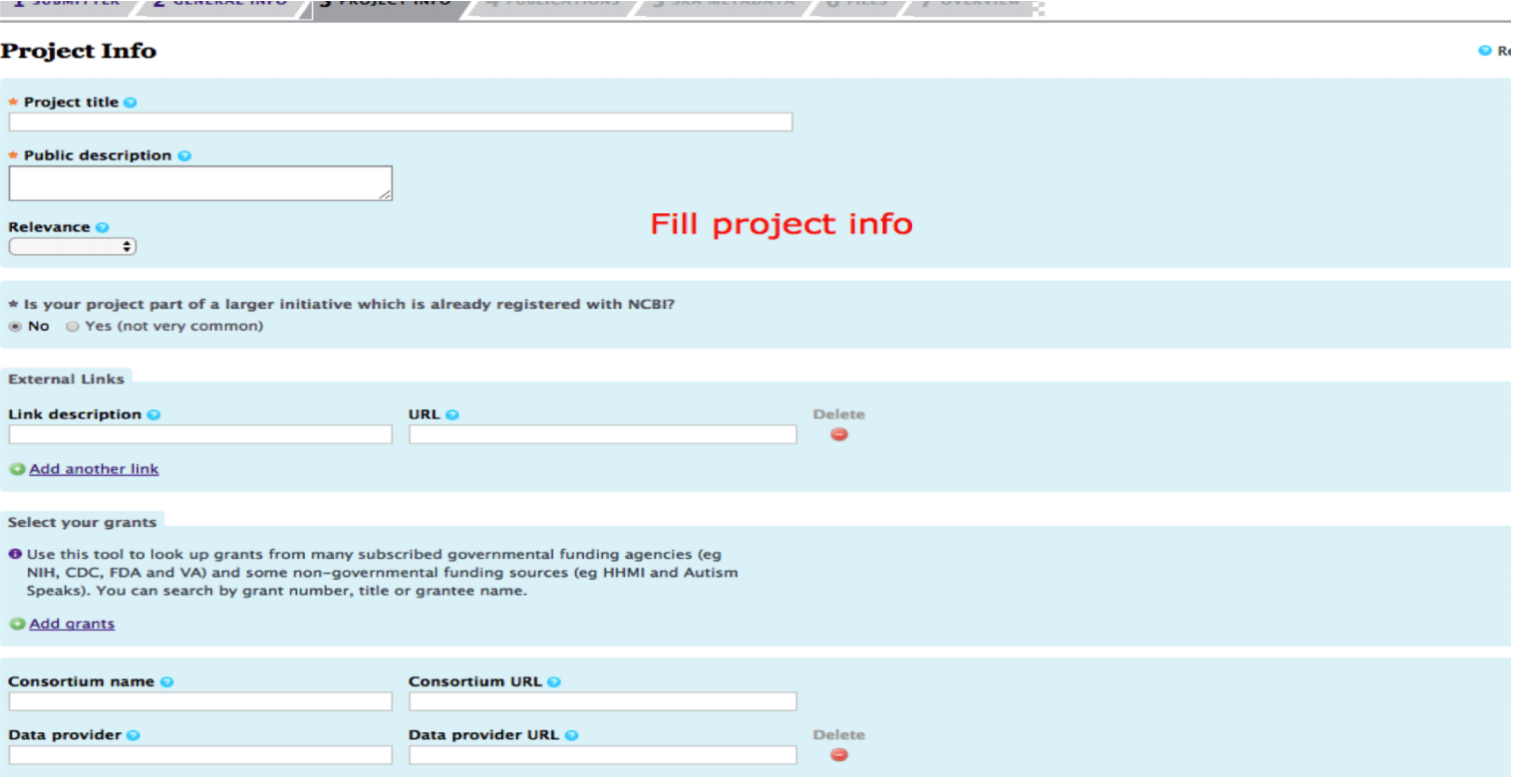
Fill in the project information. Add as much relevant information you can add in description. It will help later in searching the particular submission.
The AIRR BioSample template is not yet listed on the NCBI website. The template sheet
AIRR_BioSample_V1.0.xlscan be downloaded from airr-community/airr-standards. Fill in the required field and save the file as tab-delimited text file (.TSV format), then upload it.To submit the SRA metadata use the
AIRR_SRA_v1.0.xlsfile. Make sure that the columnsample_nameuses sample names that match the record in the BioSample template (if new BioSamples are being submitted) or a previously entered record. Also this file must be saved as tab-delimited text file for upload.Submit the raw sequence file.
Complete the submission.
Proceed with deposit of processed data, below.
Option 3. Submission via NCBI’s FTP server, using a predefined XML template#
In addition to the web interface, NCBI provides an FTP-based solution to submit bulk metadata. The corresponding AIRR XML templates can be found under airr-community/airr-standards. Otherwise users should refer to the current SRA file upload manual https://www.ncbi.nlm.nih.gov/sra/docs/submitfiles/. Users planning to frequently submit AIRR-seq data to SRA using scripts to generate the XML files MUST ensure that the templates are identical to the current upstream version on Github.
Step 2. Processed MiAIRR data submission to GenBank/TLS#
Processed sequence data will be submitted to the “Targeted Locus Study” (TLS) section of GenBank. The details of this submission process are currently still being finalized. Basically the procedure is identical to a conventional GenBank submission with the exception of additional keywords marking it as TLS submission.
Non-productive records should be removed before the data submission or use an alternative annotation as described in the specification document.
Generating MiAIRR compliant GenBank/TLS submissions: https://changeo.readthedocs.io/en/stable/examples/genbank.html
GenBank provides multiple tools (GUI and command-line) to submit data:
BankIt, a web-based submission tool with wizards to guide the submission process
Sequin, NCBI’s stand-alone submission tool with wizards to guide the submission process is available by FTP for use on for Windows, macOS and Unix platforms.
Tbl2asn is the recommended tool for the bulk data submission. It is a command-line program that automates the creation of sequence records files (.sqn) for submission to GenBank, driven by multiple tabular unput data files. Documentation and download options can be found under https://www.ncbi.nlm.nih.gov/genbank/tbl2asn2/.