ADC API Query and Analysis Example#
This example shows how repertoires and associated rearrangments may be queried from a data repository using the ADC API and then a simple analysis is performed. The example is split between two python scripts; one that performs the query and saves the data into files, and another that reads the data from the files and generates a grouped CDR3 amino acid length distribution plot. The two scripts could be combined into one, but this example illustrates how the data can be saved into files for later use. The example uses the AIRR standards python library.
Data#
This example retrieves data for the following study, which is identified by NCBI BioProject PRJNA300878. In this example, we are only going to query and retrieve the T cell repertoires.
Rubelt, F. et al., 2016. Individual heritable differences result in unique cell lymphocyte receptor repertoires of naive and antigen-experienced cells. Nature communications, 7, p.11112.
Basic study description:
5 pairs of human twins
B-cells and T-cells sequenced
B-cells sorted into naive and memory
T-cells sorted into naive CD4, naive CD8, memory CD4 and memory CD8
Total of 60 repertoires: 20 B-cell repertoires and 40 T-cell repertoires
Walkthrough#
We’ll use the airr-standards docker image for this example, which comes loaded with all the python packages needed. You will want to map a local directory inside the docker container so you can access the data and analysis results afterwards:
# Download the image
docker pull airrc/airr-standards:latest
# Make local temporary directory to hold the data
mkdir adc_example
cd adc_example
# Invoke a shell session inside the docker image
docker run -it -v $PWD:/data airrc/airr-standards:latest bash
The first python script queries the data from the VDJServer data repository and saves them into files:
# Query the data
cd /data
python3 /airr-standards/docs/examples/api/retrieve_data.py
Only a subset of the data is downloaded for illustration purposes, but
review the code to see how all data can be downloaded. A total of 40
repertoires and 300,178 rearrangements should be downloaded. The
repertoire metadata is saved in the repertoires.airr.json file,
and the rearrangements are saved in the rearrangements.tsv
file. The script should take a few minutes to run and produce the
following display messages:
Info: VDJServer Community Data Portal
version: 1.3
description: VDJServer ADC API response for repertoire query
Received 40 repertoires.
Retrieving rearrangements for repertoire: 5168912186246295065-242ac11c-0001-012
Retrieved 9768 rearrangements for repertoire: 5168912186246295065-242ac11c-0001-012
Retrieving rearrangements for repertoire: 5338391595746455065-242ac11c-0001-012
Retrieved 5521 rearrangements for repertoire: 5338391595746455065-242ac11c-0001-012
Retrieving rearrangements for repertoire: 4858300151399575065-242ac11c-0001-012
Retrieved 2885 rearrangements for repertoire: 4858300151399575065-242ac11c-0001-012
Retrieving rearrangements for repertoire: 5039977268020375065-242ac11c-0001-012
Retrieved 4053 rearrangements for repertoire: 5039977268020375065-242ac11c-0001-012
Retrieving rearrangements for repertoire: 6240077029868695065-242ac11c-0001-012
Retrieved 3506 rearrangements for repertoire: 6240077029868695065-242ac11c-0001-012
Retrieving rearrangements for repertoire: 6389112395039895065-242ac11c-0001-012
Retrieved 2289 rearrangements for repertoire: 6389112395039895065-242ac11c-0001-012
Retrieving rearrangements for repertoire: 5939858815878295065-242ac11c-0001-012
Retrieved 3637 rearrangements for repertoire: 5939858815878295065-242ac11c-0001-012
Retrieving rearrangements for repertoire: 6088937130722455065-242ac11c-0001-012
Retrieved 9028 rearrangements for repertoire: 6088937130722455065-242ac11c-0001-012
Retrieving rearrangements for repertoire: 7446748091679895065-242ac11c-0001-012
Retrieved 1540 rearrangements for repertoire: 7446748091679895065-242ac11c-0001-012
Retrieving rearrangements for repertoire: 7591789137265815065-242ac11c-0001-012
Retrieved 10000 rearrangements for repertoire: 7591789137265815065-242ac11c-0001-012
Retrieving rearrangements for repertoire: 7066128089908375065-242ac11c-0001-012
Retrieved 5662 rearrangements for repertoire: 7066128089908375065-242ac11c-0001-012
Retrieving rearrangements for repertoire: 5624006920930455065-242ac11c-0001-012
Retrieved 10000 rearrangements for repertoire: 5624006920930455065-242ac11c-0001-012
Retrieving rearrangements for repertoire: 8961797805343895065-242ac11c-0001-012
Retrieved 1179 rearrangements for repertoire: 8961797805343895065-242ac11c-0001-012
Retrieving rearrangements for repertoire: 9084118473933975065-242ac11c-0001-012
Retrieved 4464 rearrangements for repertoire: 9084118473933975065-242ac11c-0001-012
Retrieving rearrangements for repertoire: 8485700680582295065-242ac11c-0001-012
Retrieved 3908 rearrangements for repertoire: 8485700680582295065-242ac11c-0001-012
Retrieving rearrangements for repertoire: 7309695685264535065-242ac11c-0001-012
Retrieved 10000 rearrangements for repertoire: 7309695685264535065-242ac11c-0001-012
Retrieving rearrangements for repertoire: 8425807333172056551-242ac11c-0001-012
Retrieved 6863 rearrangements for repertoire: 8425807333172056551-242ac11c-0001-012
Retrieving rearrangements for repertoire: 8263242821018456551-242ac11c-0001-012
Retrieved 10000 rearrangements for repertoire: 8263242821018456551-242ac11c-0001-012
Retrieving rearrangements for repertoire: 8733756488295256551-242ac11c-0001-012
Retrieved 5298 rearrangements for repertoire: 8733756488295256551-242ac11c-0001-012
Retrieving rearrangements for repertoire: 8602072790999896551-242ac11c-0001-012
Retrieved 10000 rearrangements for repertoire: 8602072790999896551-242ac11c-0001-012
Retrieving rearrangements for repertoire: 7313153105470296551-242ac11c-0001-012
Retrieved 9121 rearrangements for repertoire: 7313153105470296551-242ac11c-0001-012
Retrieving rearrangements for repertoire: 6964444710708056551-242ac11c-0001-012
Retrieved 10000 rearrangements for repertoire: 6964444710708056551-242ac11c-0001-012
Retrieving rearrangements for repertoire: 7640859110155096551-242ac11c-0001-012
Retrieved 10000 rearrangements for repertoire: 7640859110155096551-242ac11c-0001-012
Retrieving rearrangements for repertoire: 7461458326201176551-242ac11c-0001-012
Retrieved 10000 rearrangements for repertoire: 7461458326201176551-242ac11c-0001-012
Retrieving rearrangements for repertoire: 5953881855632216551-242ac11c-0001-012
Retrieved 5916 rearrangements for repertoire: 5953881855632216551-242ac11c-0001-012
Retrieving rearrangements for repertoire: 7158276584776536551-242ac11c-0001-012
Retrieved 10000 rearrangements for repertoire: 7158276584776536551-242ac11c-0001-012
Retrieving rearrangements for repertoire: 6393557657723736551-242ac11c-0001-012
Retrieved 7257 rearrangements for repertoire: 6393557657723736551-242ac11c-0001-012
Retrieving rearrangements for repertoire: 6205695788196696551-242ac11c-0001-012
Retrieved 10000 rearrangements for repertoire: 6205695788196696551-242ac11c-0001-012
Retrieving rearrangements for repertoire: 4476756703191896551-242ac11c-0001-012
Retrieved 10000 rearrangements for repertoire: 4476756703191896551-242ac11c-0001-012
Retrieving rearrangements for repertoire: 4357957907784536551-242ac11c-0001-012
Retrieved 7033 rearrangements for repertoire: 4357957907784536551-242ac11c-0001-012
Retrieving rearrangements for repertoire: 4931851437876056551-242ac11c-0001-012
Retrieved 10000 rearrangements for repertoire: 4931851437876056551-242ac11c-0001-012
Retrieving rearrangements for repertoire: 4744762662462296551-242ac11c-0001-012
Retrieved 10000 rearrangements for repertoire: 4744762662462296551-242ac11c-0001-012
Retrieving rearrangements for repertoire: 3252733973504856551-242ac11c-0001-012
Retrieved 10000 rearrangements for repertoire: 3252733973504856551-242ac11c-0001-012
Retrieving rearrangements for repertoire: 2989624276951896551-242ac11c-0001-012
Retrieved 10000 rearrangements for repertoire: 2989624276951896551-242ac11c-0001-012
Retrieving rearrangements for repertoire: 3628844259615576551-242ac11c-0001-012
Retrieved 5208 rearrangements for repertoire: 3628844259615576551-242ac11c-0001-012
Retrieving rearrangements for repertoire: 3438706057421656551-242ac11c-0001-012
Retrieved 9530 rearrangements for repertoire: 3438706057421656551-242ac11c-0001-012
Retrieving rearrangements for repertoire: 2197374609531736551-242ac11c-0001-012
Retrieved 10000 rearrangements for repertoire: 2197374609531736551-242ac11c-0001-012
Retrieving rearrangements for repertoire: 1993707260355416551-242ac11c-0001-012
Retrieved 10000 rearrangements for repertoire: 1993707260355416551-242ac11c-0001-012
Retrieving rearrangements for repertoire: 2541616238306136551-242ac11c-0001-012
Retrieved 6512 rearrangements for repertoire: 2541616238306136551-242ac11c-0001-012
Retrieving rearrangements for repertoire: 2366080924918616551-242ac11c-0001-012
Retrieved 10000 rearrangements for repertoire: 2366080924918616551-242ac11c-0001-012
The second python script loads the data from the files and generates a CDR3 amino acid length distribution that is grouped by the T cell subset. This study performs flow sorting to generate four T cell subsets: naive CD4+, naive CD8+, memory CD4+, memory CD8+. The script uses the repertoire metadata to determine the T cell subset for the rearrangement, tabulates the counts, normalizes them, and generates a grouped bar chart with the results:
# Run the analysis
python3 /airr-standards/docs/examples/api/analyze_data.py
The figure is placed in the plot.png file and should look like this:
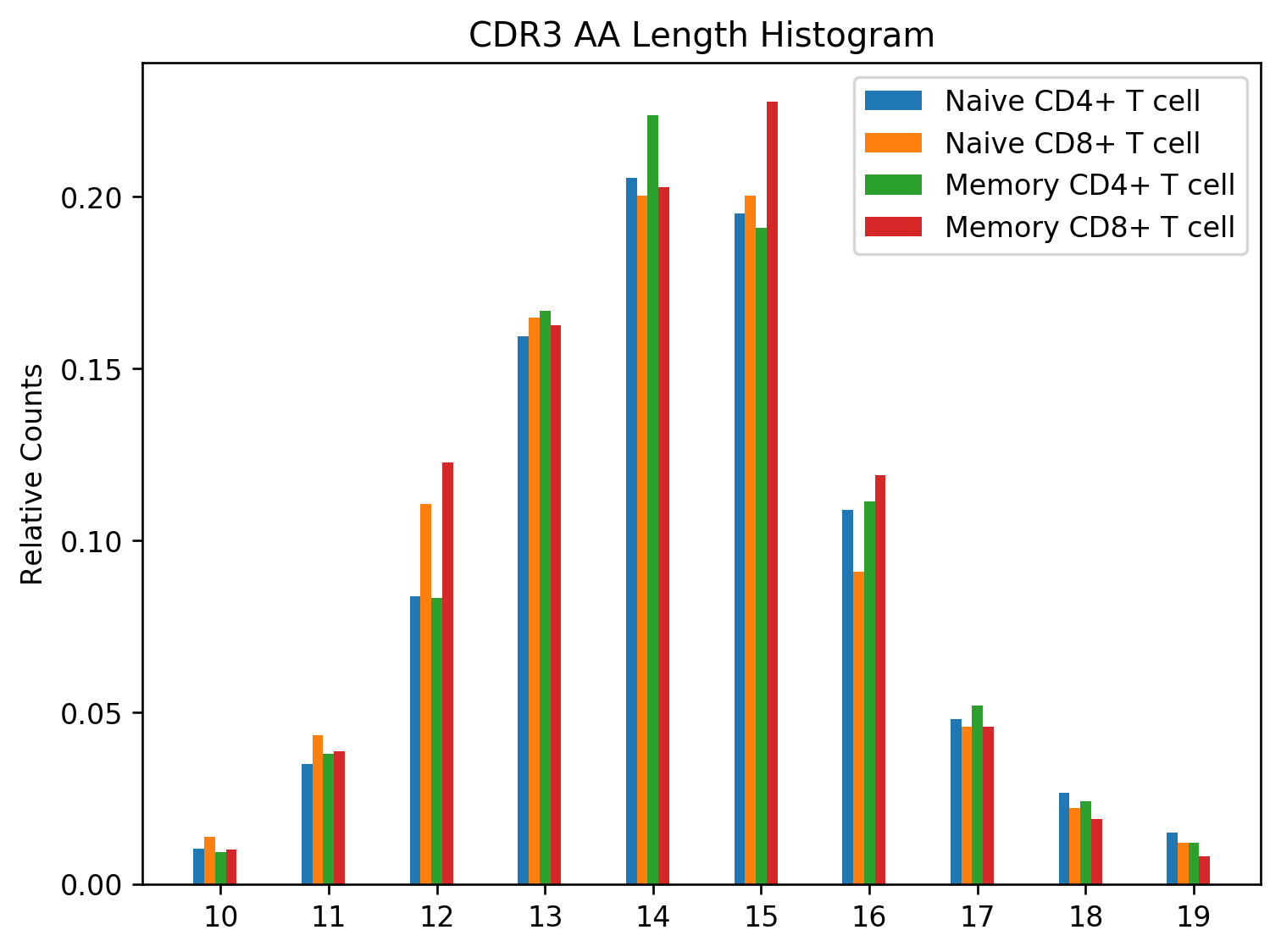
CDR3 AA Length Histogram grouped by T cell subsets.#